The DNA assembly challenge
Simply put, the DNA assembly challenge is to take a set of double-stranded DNA sequence fragments, and stitch them together in the proper order and proper orientation to yield a single, potentially circular, assembled DNA sequence. These DNA sequence fragments are often referred to as "parts" in the synthetic biology lexicon, especially when the fragments are each associated with a particular biological function, such as a promoter, a coding sequence, a terminator, etc. Here in this manual, "parts" simply refer to generalized DNA sequences.
Here is an example DNA assembly:
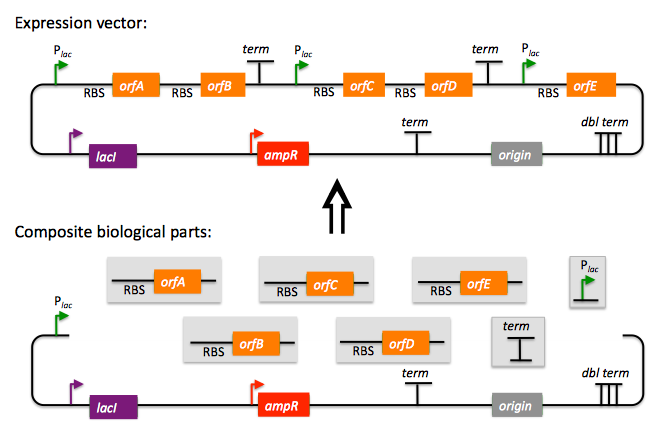
We start with 8 non-degenerate (non-repeated) composite biological parts (shown at the bottom of the figure): a vector backbone, 5 protein coding sequences (orfA to orfE) with upstream ribosome binding sites (RBS), a terminator, and a promoter. We assemble these 8 parts into the target expression vector (shown at the top of the figure). Note that in this particular example, we used the same terminator and promoter parts twice to achieve the target expression vector.
For a recent review on the many available experimental methodologies for addressing this challenge, see (Hillson 2011, Ellis 2011). The following sections of this introduction will specifically focus on the traditional, BioBrick, SLIC/Gibson/CPEC/SLiCE, and Golden Gate approaches.
